COMPUTATIONAL CHEMOGENOMICS FOR DRUG ABUSE (CC4DA)
PI: Xiang-Qun (Sean) Xie, PhD, MBA

The Computational Chemogenomics Core for Drug Abuse (CC4DA, or Core A) of CDAR, is proposed specifically to address existing fundamental challenges of drug abuse (DA) and medication research to enhance the NIH/NIDA funded research projects (FRP), with particular attention to polydrug addiction and polypharmacology, via systematic investigation of the interaction between chemical compounds and DA-related protein targets, as well as the underlying signaling pathways. We will build upon our existing chemogenomics and computational expertise in the realm of cannabinoid research and expand our developed computational algorithms and tools to facilitate research in the general domain of DA. To accomplish the proposed objectives, the CC4DA Core will: (i) enable data sharing and processing among scientists in the DA and related scientific communities by our established public chemogenomics knowledgebase for drug abuse (DA-KB); (ii) integrate and further advance our established computational algorithms and tools for exploration and prediction of DA targets, DA pathways, the underlying mechanisms, and the potential polypharmacological effects that relate to polydrug addiction; and (iii) implement cloud computing and sourcing DA research services (CloudDA) to facilitate in silico design, discovery and development of new medications with potential for DA treatment. Strategies used to complete the proposed work will overcome knowledge barriers of advanced computer modeling, complicated machine learning methods and high-performance computing for scientists/users in the fields of pharmaceutical, neurobiological and pharmacological sciences. Successful development of DA-KB and computational algorithms for DA research (DAR) will provide powerful tools for analyzing a wide variety of chemicals, and effectively assisting scientists in the rational design of DA therapeutics.
Current Research
Establishment of cloud computing drug-abuse and neurobiological diseases domain-specific Chemogenomics databases for drug abuse (DA) related research.
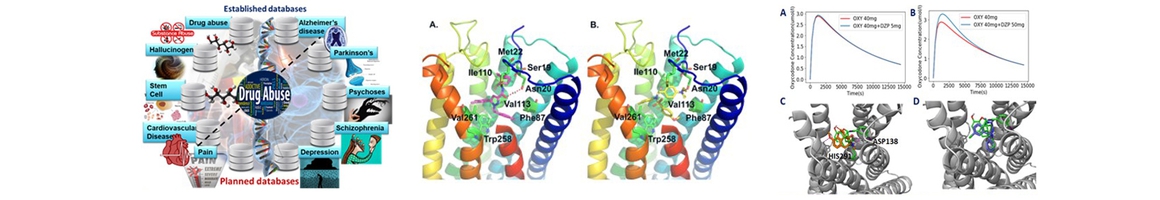
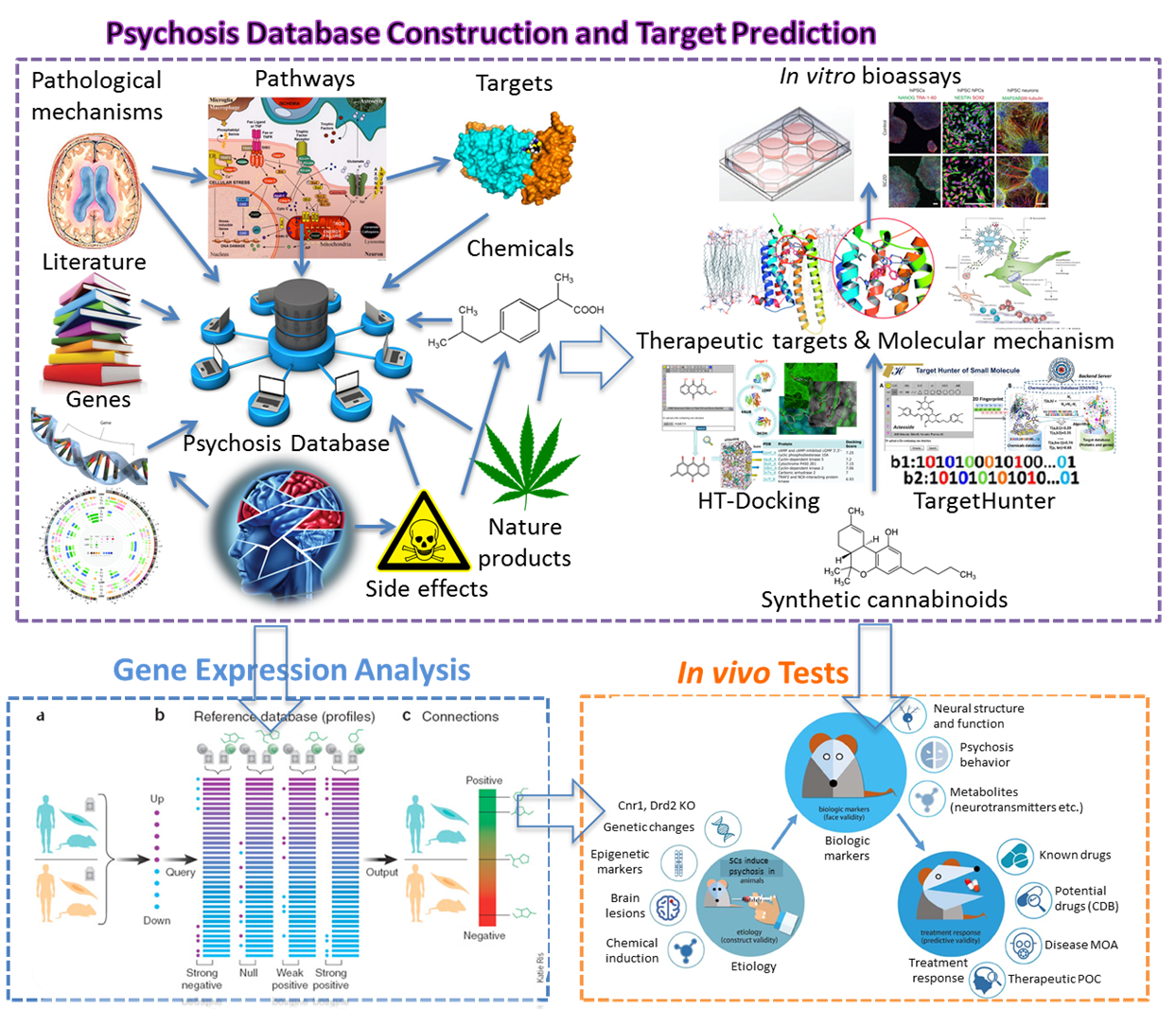
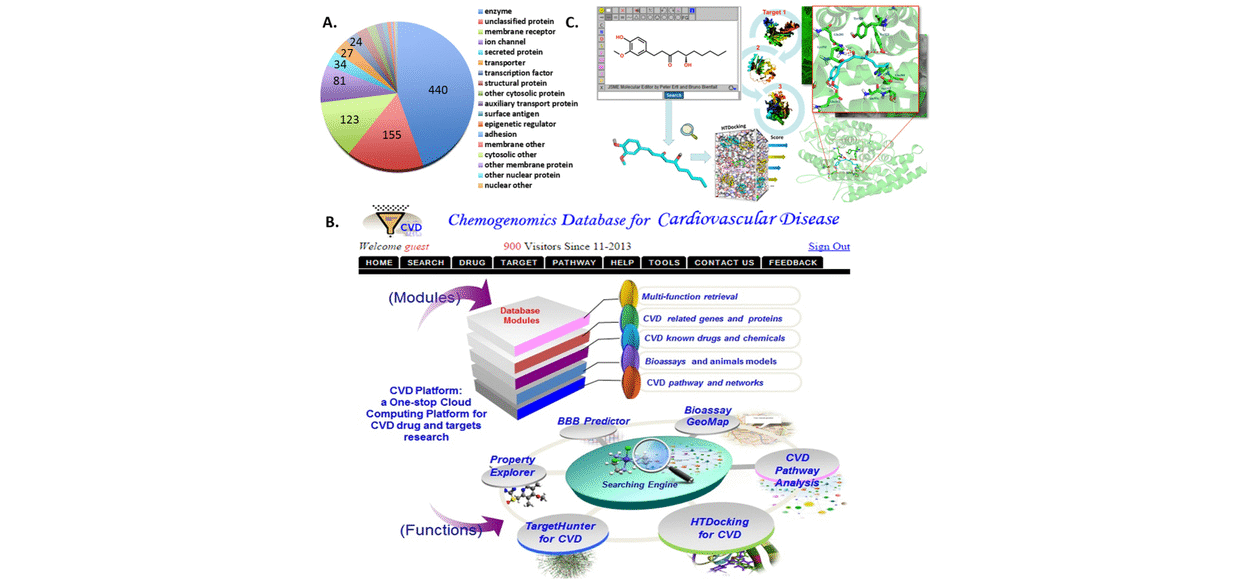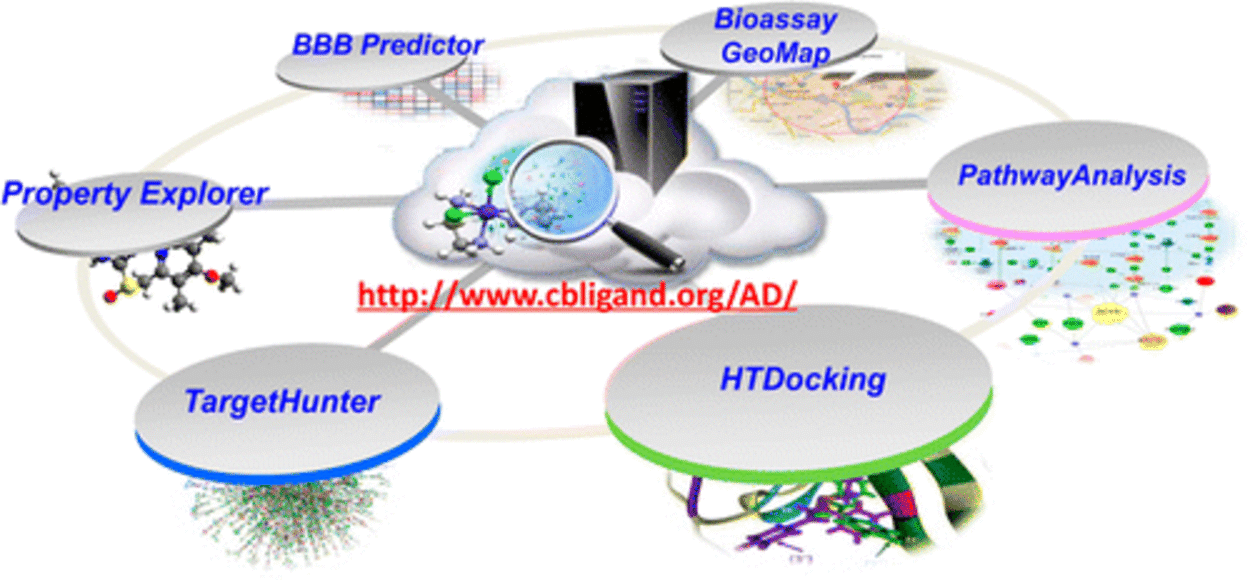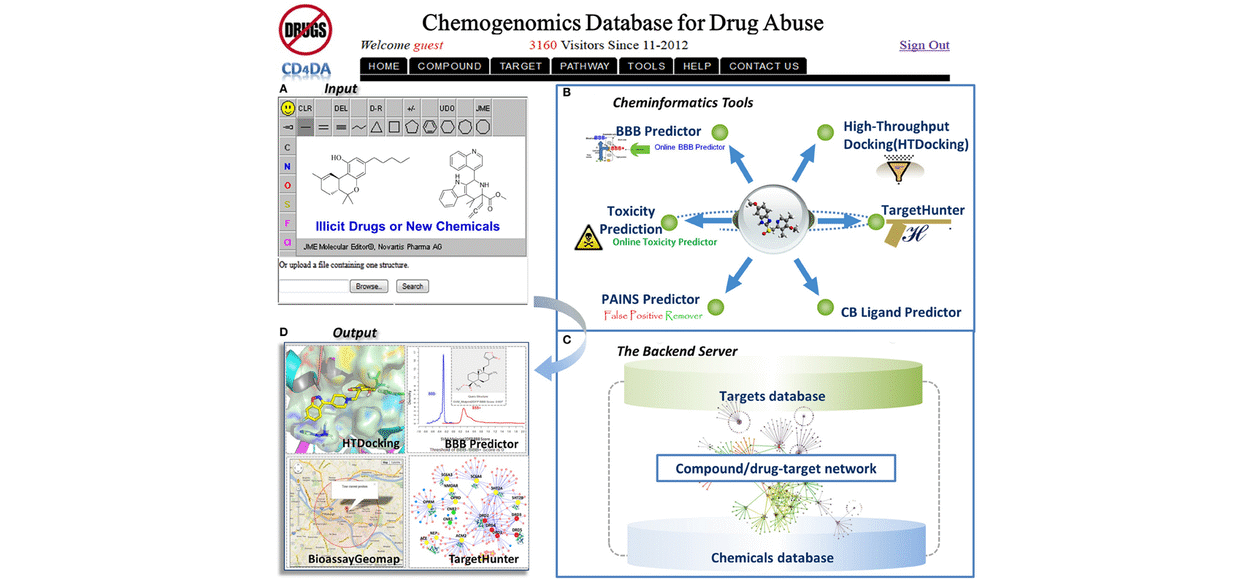 We have collected disease-specific genes, proteins, signal pathways and small molecules to build chemogenomics databases, including drug abuse, hallucinogen, Alzheimer’s disease, cardiovascular disease and stem cell. These platforms are integrated with online structure search function and data analysis tools to facilitate data-sharing and information exchange among scientific research communities for target/off-target identification neurbiological drug abuse research. CNS drugs and their corresponding target proteins and pathways have been data mined to enable the system pharmacology investigation of possible side effects, with the understanding that the observed phenotypes may originate from direct or indirect interactions with targets relevant to DA.
We have collected disease-specific genes, proteins, signal pathways and small molecules to build chemogenomics databases, including drug abuse, hallucinogen, Alzheimer’s disease, cardiovascular disease and stem cell. These platforms are integrated with online structure search function and data analysis tools to facilitate data-sharing and information exchange among scientific research communities for target/off-target identification neurbiological drug abuse research. CNS drugs and their corresponding target proteins and pathways have been data mined to enable the system pharmacology investigation of possible side effects, with the understanding that the observed phenotypes may originate from direct or indirect interactions with targets relevant to DA.
Development of GPU-acceleraed computational chemogenomics algorithms/tools for target identification to study the poly-pharmacophore and polyaddiction of abused drugs and neurodisorders.
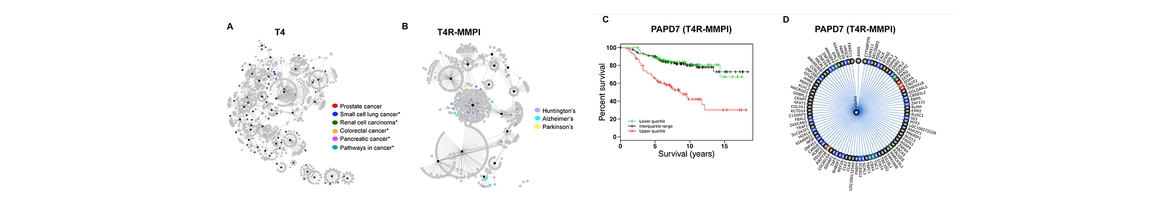
Target identification is one of the most important steps to study the structure and mechanism of diverse abused drugs. We have developed TargetHunter and HTDocking algorithms to predict potential protein targets of small molecules based on ligand structure and protein structures through molecular similarity and molecular docking simulation, respectively. (1) We have built a user-friendly interface and cytoscape plot for result visualization; (2) We have designed Proselection algorithm to select subsets of protein structures from multiple instances of an individual protein target with better capability of distinguishing active and inactive ligands; and (3) We have further advanced our established computational algorithms and tools by applying both HTDocking and TargetHunter for exploration and prediction of DA targets, DA pathways, and the underlying mechanisms and potential polypharmacological effects of 5F-AMB/5F-ADB.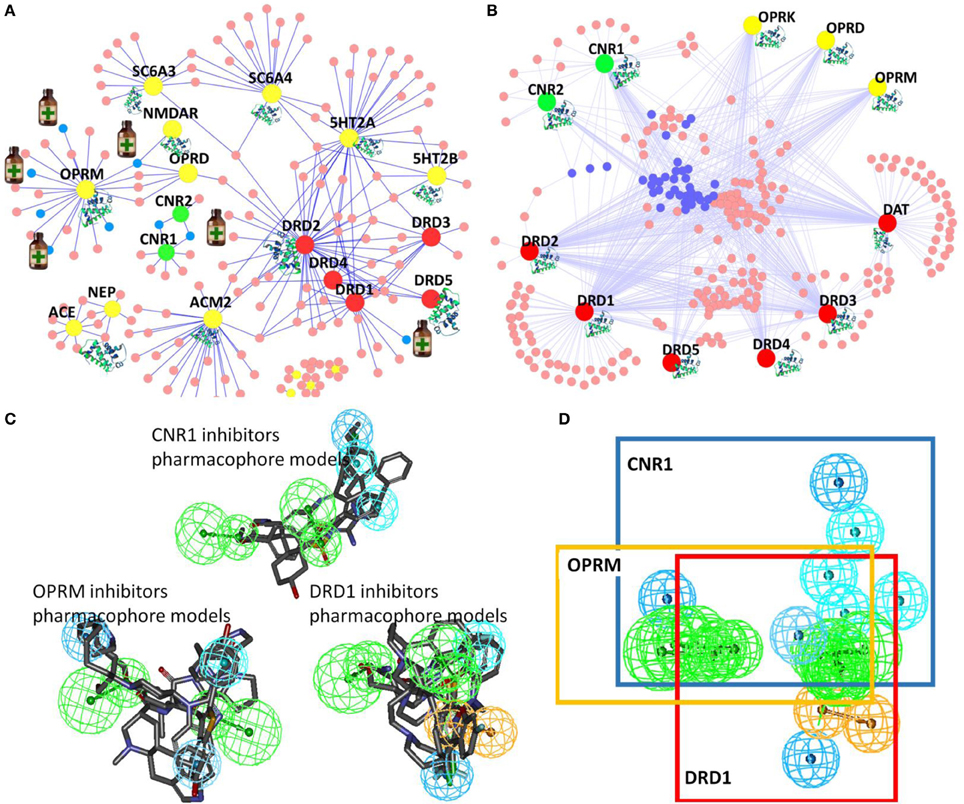
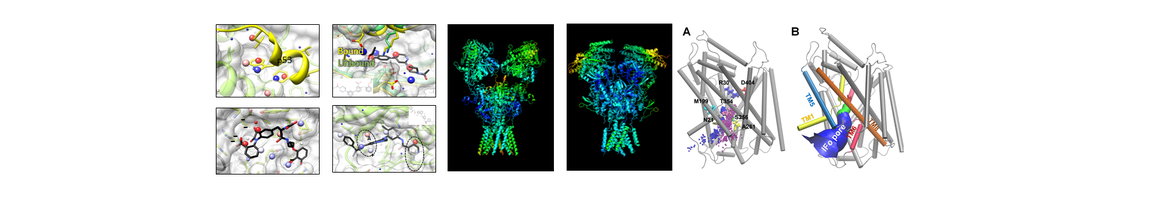
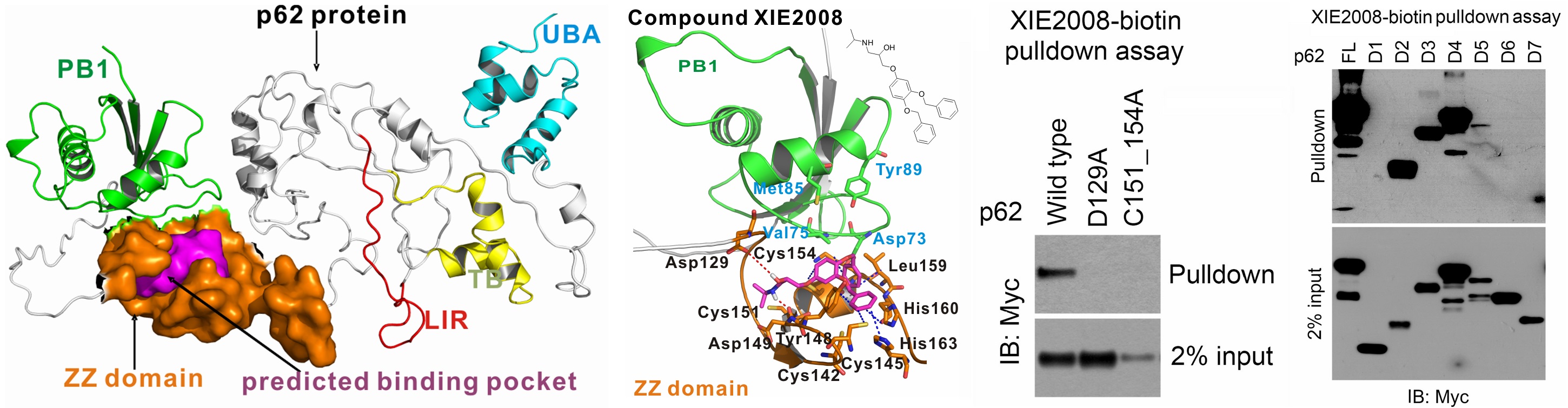
Advancement and Applications of machine-/deep- learning computing chemogenomics algorithmstools for design of novel small molecular probes for drug abuse and neurodisorders related research. We have created online tools and implemented cloud computing/sourcing DA research services to facilitate in silico design, discovery and development of new medications for treatment of DA and neuro-diseases. (1) The ML-guided blood-brain-barrier (BBB) predictor provides service for predicting the capability of brain penetration for small molecules; (2) The PAINS remover and neuro-toxicity server are designed to pan assay interference compounds and toxic chemicals, respectively; (3) we have developed a prototype of fragment-based approach for in silico allosteric ligand design and have successfully applied it in the design of mGlu5 ligands; and (4) we have applied our computational chemogenomics tools to design small molecular probes of p62, a key player in autophagy process with the potential for treatment of Huntington disease.
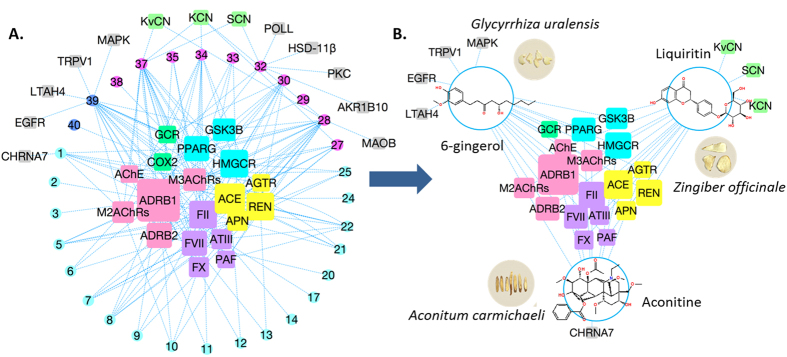
Applications of our systems pharmacology approaches to associate molecular mechanisms with clinical outcome. In order to understand the molecular mechanism(s) behind the effects of medications on patient outcomes and the potential synergisms in clinical contexts, we applied molecular systems pharmacology (MSP) techniques based on our established chemogenomic databases. MSP applies the principles of systems biology to study pharmacology, and it seeks to understand how medicines work on various molecular targets from different systems of the body. (1) We have simulated the drug-drug interactions between Oxycodone and Diazepam to quantitatively assess the toxicity effects; (2) we have analyzed the synergistic effects of anti-hypertensive drugs and cholinesterase inhibitors on cognitive decline in AD patients; (3) we have investigated drug synergy mechanisms of an herbal formula for cardiovascular disease; and (4) we have conducted network system pharmacology analysis on aspirin/cilostazol to study their therapeutic effects on cardiovascular disease associated brain CNS.
New collaborations for drug abuse and neurodisorder related research. We have established collaborations with investigators from local and international Universities, centers of drug abuse and neurodisorder for drug abuse and other neurological diseases. (1) We have worked with Dr. Mary Abood from Temple University, NIDA P30 center to study the molecular mechanism of synthetic cannabinoids on chronic pain; (2) In collaboration with Dr. Bahar’s lab (Core B) and Dr. Yigong Shi (Tsinghua University), we have determined the mechanism of 𝛾-secretase function, as well as its allosteric modulation or inhibition by substrate binding. (3) We are also working with Dr. Eric Xing and Dr. Wei Wu (Core C) to identify small molecules that can reverse the gene signatures induced by abused drugs for potential DA treatment; and (4) we initiated collaboration with Dr. Robert Sweet for cannabinoid-induced psychosis.
References
- Xiang-Qun Xie,* Lirong Wang, Haibin Liu, Qin Ouyang, Cheng Fang, and Weiwei Su.(2014) Chemogenomics knowledgebased polypharmacology analyses of drug abuse related G-protein coupled receptors and their ligands. Front Pharmacol. doi: 10.3389/fphar.2014.00003
- Liu, H., Wang, L., Su, W., & Xie, X. Q. (2014). Advances in recent patent and clinical trial drug development for Alzheimer’s disease. Pharmaceutical patent analyst, 3(4), 429-447.
- Xu X, Ma S, Feng Z, Hu G, Wang L, Xie, X.-Q*. Chemogenomics knowledgebase and systems pharmacology for hallucinogen target identification-Salvinorin A as a case study. J Mol Graph Model. 2016 Nov;70:284-295. PMID: 27810775
- Zhang H, Ma S, Feng Z, Wang D, Li C, Cao Y, Chen X, Liu A, Zhu Z, Zhang J, Zhang G, Chai Y, Wang L, Xie, X.-Q*. Cardiovascular Disease Chemogenomics Knowledgebase-guided Target Identification and Drug Synergy Mechanism Study of an Herbal Formula. Sci Rep. 2016 Sep 28;6:33963. PMID: 27678063
- Zhang Y, Wang L, Feng Z, Cheng H, McGuire TF, Ding Y, Cheng T, Gao Y, Xie, X.-Q*. StemCellCKB: An Integrated Stem Cell-Specific Chemogenomics KnowledgeBase for Target Identification and Systems-Pharmacology Research. J Chem Inf Model. 2016 Oct 24;56(10):1995-2004. PMID: 27643925
- Lirong Wang, Chao Ma, Peter Wipf, Haibin Liu, Weiwei Su, Xiang-Qun Xie. TargetHunter: An In Silico Target Identification Tool for Predicting Therapeutic Potential of Small Organic Molecules Based on Chemogenomic Database. AAPS J. 2013 Apr; 15(2): 395–406. Published online 2013 Jan 5. doi: 10.1208/s12248-012-9449-z
- Bian Y, Feng Z, Yang P, Xie, X.-Q*. Integrated In Silico Fragment-Based Drug Design: Case Study with Allosteric Modulators on Metabotropic Glutamate Receptor 5. AAPS J. 2017 Jul;19(4):1235-1248.
- Cha-Molstad H, Yu JE, Feng Z, Lee SH, Kim JG, Yang P, Han B, Sung KW, Yoo YD, Hwang J, McGuire T, Shim SM, Song HD, Ganipisetti S, Wang N, Jang JM, Lee MJ, Kim SJ, Lee KH, Hong JT, Ciechanover A, Mook-Jung I, Kim KP, Xie, X.-Q*, Kwon YT*, Kim BY*. p62/SQSTM1/Sequestosome-1 is an N-recognin of the N-end rule pathway which modulates autophagosome biogenesis. Nature Communications 8, Article number: 102 (2017).
- Ying Xue et al. The efficacy and safety of cilostazol as an alternative to aspirin after coronary stent implantation in patients with aspirin intolerance: a combination clinical study and computational system pharmacology analysis of cilostazol as an alternative to aspirin after PCI. Acta Pharmacologica Sinica, 2017, in press
- Feng, Z.; Pearce, L. V.; Xu, X.; Yang, X.; Yang, P.; Blumberg, P. M.; Xie, X.-Q. Structural Insight into Tetrameric hTRPV1 from Homology Modeling, Molecular Docking, Molecular Dynamics Simulation, Virtual Screening and Bioassay Validations. J. Chem. Inf. Model. 2015, In press.